mapcomp performs a spatial pattern analysis based on the calculation
of a formal distance (the Hellinger distance) between the density map of
count or incidence data, and the density map of sampling effort. Statistical
tests of spatial homogeneity are based on permutations across sampling sites
and on valuable properties of the Hellinger distance.
Usage
mapcomp(data, ...)
# S3 method for data.frame
mapcomp(
data,
delta,
bandwidth,
nperm = 100,
edge_correction = FALSE,
threads = 1,
verbose = TRUE,
...
)
# S3 method for matrix
mapcomp(
data,
delta,
bandwidth,
nperm = 100,
edge_correction = FALSE,
threads = 1,
verbose = TRUE,
...
)
# S3 method for count
mapcomp(
data,
delta,
bandwidth,
nperm = 100,
edge_correction = FALSE,
threads = 1,
verbose = TRUE,
...
)
# S3 method for incidence
mapcomp(
data,
delta,
bandwidth,
nperm = 100,
edge_correction = FALSE,
threads = 1,
verbose = TRUE,
...
)Arguments
- data
A data frame or a matrix with only three columns: the two first ones must be the x and y coordinates of the sampling units, and the last one, the corresponding disease intensity observations. It can also be a
countor anincidenceobject.- ...
Additional arguments to be passed to other methods.
- delta
Mesh size of the grid over the geographical domain of the sampling units used to compute the integral Hellinger distance between the probability density function of observations and the probability density function of sampling effort.
- bandwidth
Bandwidth parameter for smoothing. It allows to test the spatial extent of heterogeneity if any.
- nperm
Number of random permutations to assess probabilities.
- edge_correction
Apply edge correction to account for the fact that bordering points intrinsically suffer from a lack of neighboring observation sites. FALSE by default.
- threads
Number of threads to perform the computations.
- verbose
Explain what is being done (TRUE by default).
Value
An object of class mapcomp, which is a list containing the following
components:
data | The input data. |
coord | The coordinates and normalized intensity for each point of the full grid. |
object | The class of data. |
bandwidth | The bandwidth parameter. |
stat, pval | The statistic and corresponding p-value (see references for more details). |
References
Lavigne C, Ricci B, Franck P, Senoussi R. 2010. Spatial analyses of ecological count data: A density map comparison approach. Basic and Applied Ecology. 11:734–742.
Examples
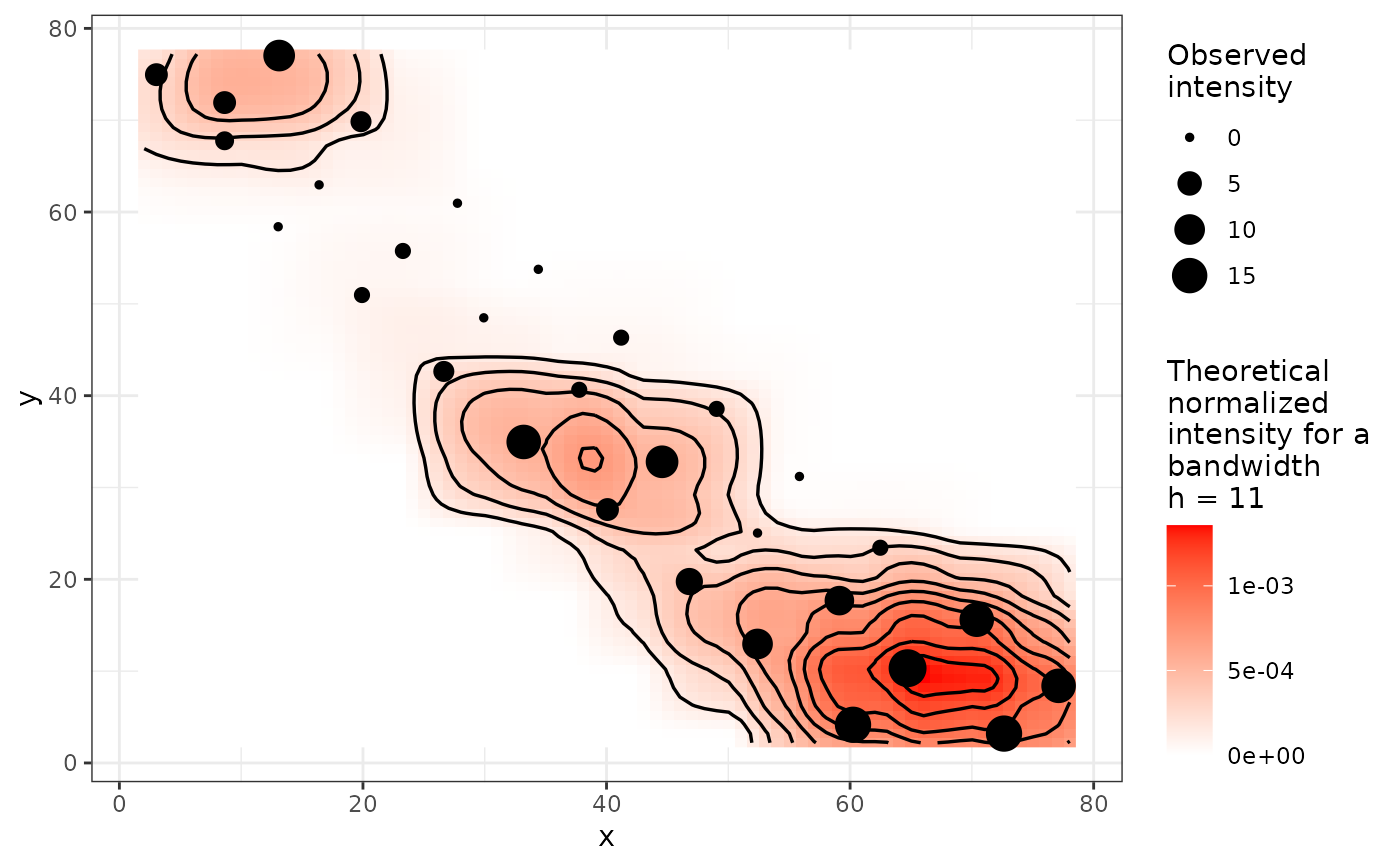
set.seed(123)
my_res <- mapcomp(codling_moths, delta = 1, bandwidth = 11,
edge_correction = FALSE, nperm = 20)
my_res
#> Map Comparison analysis (mapcomp)
#>
#> Call:
#> mapcomp.data.frame(data = codling_moths, delta = 1, bandwidth = 11,
#> nperm = 20, edge_correction = FALSE)
#>
#> Stat: 0.3423 (P = 0.047619)
#>
plot(my_res)
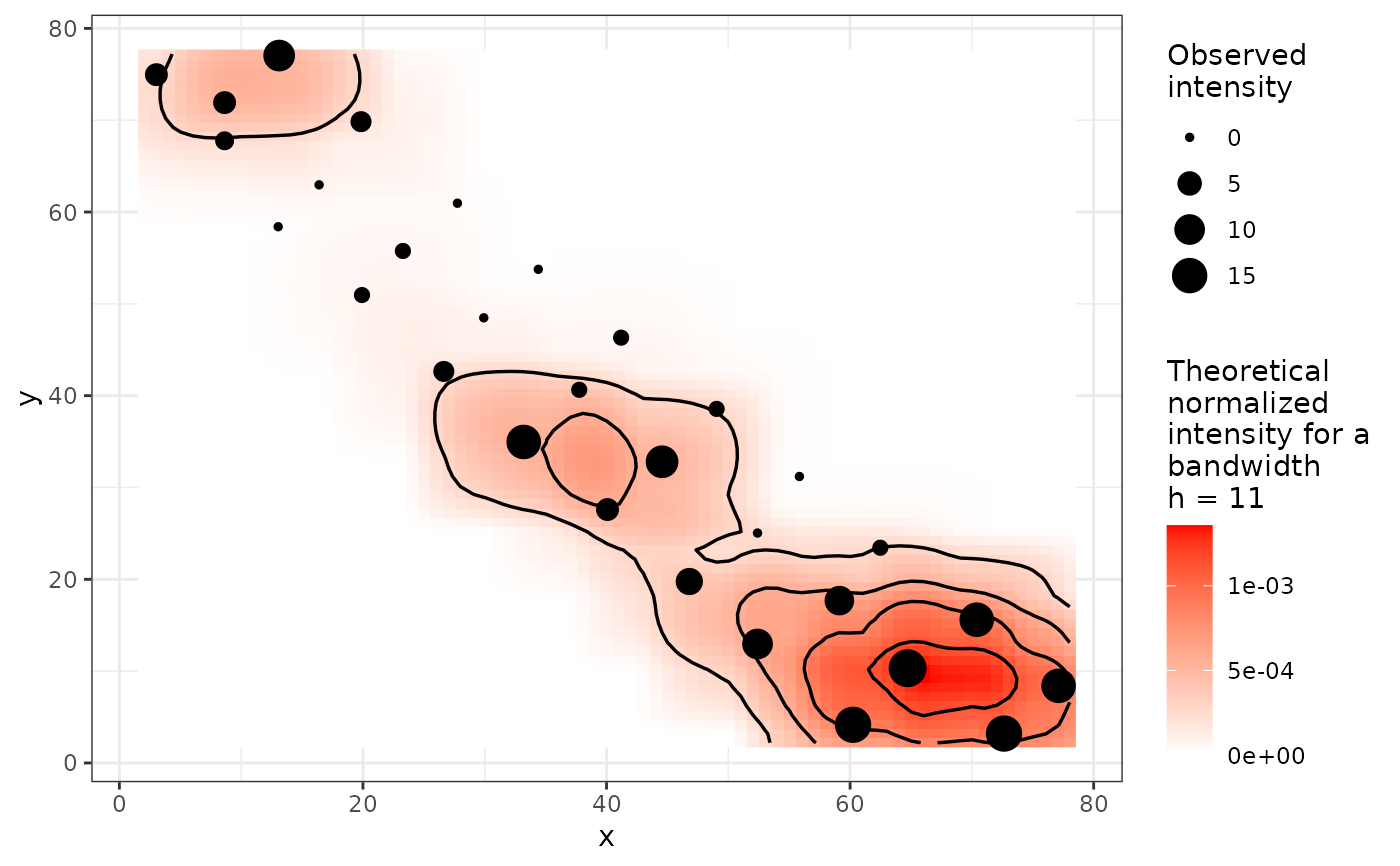 set.seed(123)
my_count <- count(codling_moths, mapping(x = xm, y = ym))
my_res <- mapcomp(my_count, delta = 1, bandwidth = 11,
edge_correction = FALSE, nperm = 20)
my_res
#> Map Comparison analysis (mapcomp)
#>
#> Call:
#> mapcomp.count(data = my_count, delta = 1, bandwidth = 11, nperm = 20,
#> edge_correction = FALSE)
#>
#> Stat: 0.3423 (P = 0.047619)
#>
plot(my_res, bins = 10)
set.seed(123)
my_count <- count(codling_moths, mapping(x = xm, y = ym))
my_res <- mapcomp(my_count, delta = 1, bandwidth = 11,
edge_correction = FALSE, nperm = 20)
my_res
#> Map Comparison analysis (mapcomp)
#>
#> Call:
#> mapcomp.count(data = my_count, delta = 1, bandwidth = 11, nperm = 20,
#> edge_correction = FALSE)
#>
#> Stat: 0.3423 (P = 0.047619)
#>
plot(my_res, bins = 10)